Server use case
Input data
Input data is the protein tri-dimensional structure. SA-Mot accepts as input either the PDB formatted coordinate file (A) or the PDB code(B) of the protein target. Input PDB files can contain several chains.
Encoding output
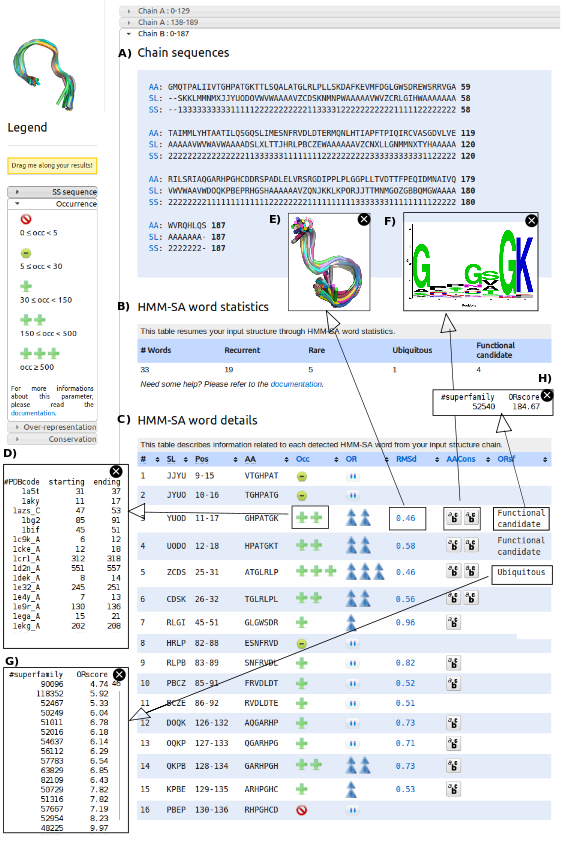
When a protein structure is uploaded, SA-Mot server proceeds for all chains contained in the uploaded structures of all SA-Ws, and the results are presented separately for each chain. The result of this process is organized in three parts. At first, SA-Mot presents the protein chain (C) using the different sequences corresponding to the primary sequence (AA), secondary-structure (SS) and three dimensional structure through the structural-letter sequence (SL). These sequences allow the users to easily identify the loop regions of the studied chain.
Then, SA-Mot provides a table containing the counts of extracted SA-Ws of interest (D). This table gives an overview of isolated SA-Ws of interest. We can observe that the protein 2RHM chain B contains 19 recurrent SA-Ws , one ubiquitous SA-W and four functional candidat words. We can already conclude that this protein contains regions of interest.
Lastly, SA-Mot provides a second interactive table allowing the identification of SA-Ws of interest (E) using statistic, geometric and sequence parameters. This interactive table contains, for each SA-W (column SW} its positions and amino-acid sequence (columns Pos and AA. Other columns contain the values of parameters used for the identification of SA-Ws of interest presented in ``SA-Mot Method'' page. Thus the columns Occ and OR presenting the occurrence and the over-representation score computed in loop dataset, allow users to identify encoded into recurrent and non random SA-Ws corresponding to structural motifs involved in the structural redundancy of loops. To illustrate the occurrence of a SA-W, users can access to the list of proteins (and positions) it has been identified in (F) by clicking on the related icon. The columns RMSd}} and AACons, corresponding to the RMSd and Zmax allow users to identify SA-Ws with a relevant structural or sequence conservation. These conservations are illustrated by figures obtained by clicking on the corresponding values. The first figure (G) corresponds to the superimposition of all fragments encoded into the SA-W and the second (H)corresponds to the logo logo of the amino-acid sequences of all fragments encoded into a SA-W. Lastly, using the column ORsf corresponding to the result of the computation of the SA-W over-representation in SCOP superfamilies, the user can locate structural motifs, which are likely involved in protein function (functional candidate words) or in protein structures (ubiquitous SA-Ws). To help users to identify the role of these SMs, they have access to the SCOP id of each superfamily where the SA-W is over-represented (I,J).
In the table SA-Ws are ranked according to their positions in the studied chain. The table can be sorted according to the different columns in order to facilitate the identification of SA-Ws of interest.
Thus, SA-Mot learn us that the region at position 130-141 is
composed of rare SA-Ws suggesting that it is flexible. Moreover,
we learn that 19 SA-Ws are recurrent and over-represented in the
loop data set. Most of the SA-Ws present weak structural
variability and amino-acid specificities, suggesting that they
correspond to structural motifs involved in crucial region in
protein.
Thus, we can conclude that regions located at positions 11-18,
25-32, 45-51, 128-135 seems to be crucial for the
protein. Moreover, we have more information about SA-Ws ZCDS,
YUOD, UODO, CDSK, OZGB. In fact SA-W ZCDS is an
ubiquitous words, suggesting that this region is involved in
protein stability and folding.
For four SA-Ws YUOD, UODO, CDSK, OZGB a
putative function role is assigned because they are strongly
over-represented in few SCOP superfamilies. YUOD
and UODO are over-represented in the superfamily
"P-loop containing nucleotide triphosphate hydrolases"
(SCOPid=52540, as presented in J) that groups protein
binding nucleotides. These results suggest the structural motifs
encoded into these SA-Ws contain residues involved in the
nucleotide-binding site. CDSK is strongly
over-represented in the superfamily ``YVTN
repeat-like/Quinoprotein amine dehydrogenase'' (SCOP id=50969)
and OZGB is the ``Trypsin-like serine proteases''
superfamily (SCOP id=50494). We can suppose that the structural
motifs encoded into OZGB is involved in the
functional site of trypsin proteins. For the three other chains,
we find also interesting SM encoded into YUOD,
UODO and OZGB.
Thus we can conclude that SA-Mot allows us an easily and rapid identification of SMs of interest for protein function. After this analysis, we can propose that each chain of protein 2RHM contains a nucleotide-binding sites at position 11-18 and a second potential functional site in position 165-171. Thus contrary to methods based on the sequence, SA-Mot allows the identification and location of potential functional sites in the uncharacterized protein 2RHM, that could help the determination of its function.